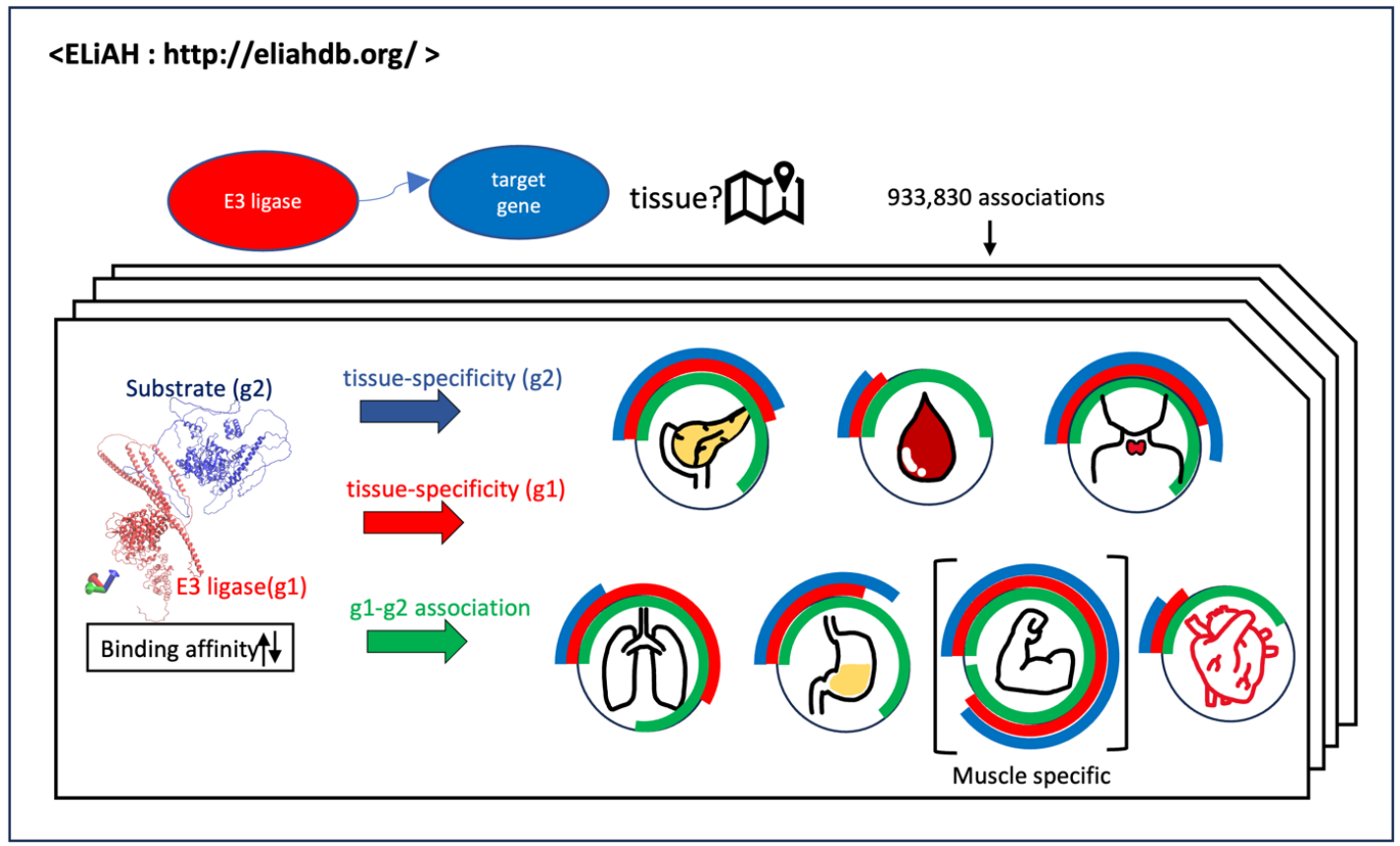
ELiAH: the atlas of E3 ligases in human tissues for targeted protein degradation
Ubiquitination-based targeted degradation of protein (TPD) including PROTAC (proteolysis-targeting chimera) has emerged as a promising pharmaceutical approach. Although the relationship between E3 ligands co-existing genes across human tissues is indispensable to prevent off-target effects, previous attempts have been mainly focused on the physicochemical properties of TPD molecules. Here, we developed E3 Ligase Atlas of Human (ELiAH; https://eliahdb.org/), a web-resource that allows us to prioritize target pairs of TPD consisting of inferred E3 ligase-target gene pairs under tissue-specific expression profiles. Leveraging over 2,900 RNA-seq profiles consisting of eleven human tissues from the Genotype-Tissue Expression (GTEx) consortium, tissue-specific genes are presented (FDR p-value of Mann-Whitney test < 0.05). Then, we identified 933,830 relationships consisting of 614 E3 ligases and 20,924 expressed genes considering the degree of tissue specificity. To further facilitate the analysis of TPD, docking properties of those relationships are also modeled using RosettaDock. Therefore, ELiAH presents a comprehensive repertoire of E3 ligases for ubiquitination-based TPD drug development, including mutually associated target genes and tissue-specific expression.